Introduction
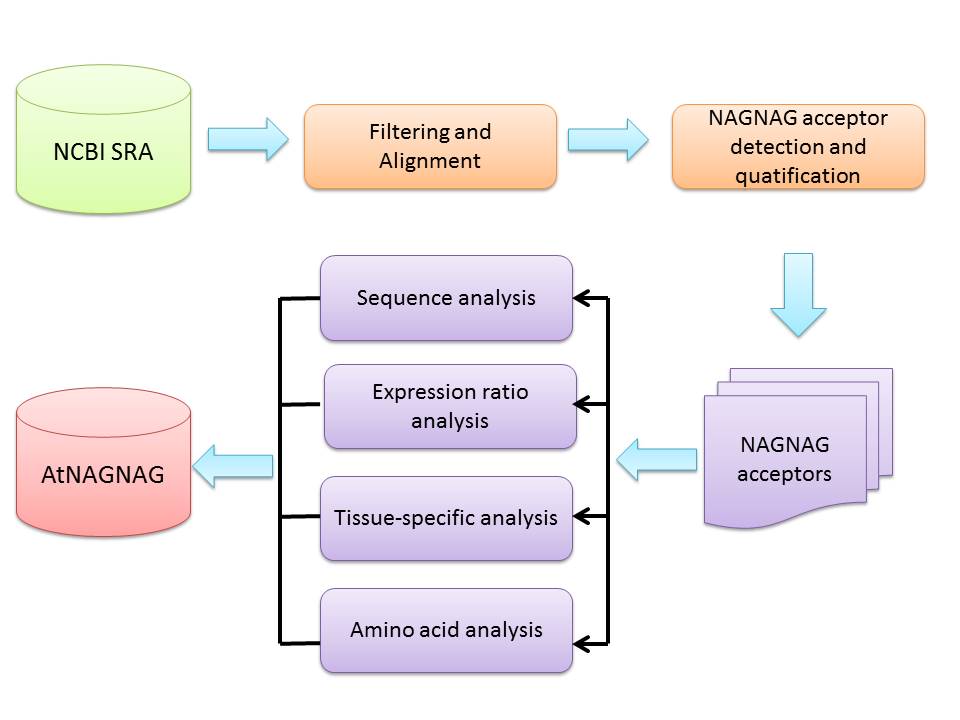
NAGNAG alternative splicing is widely present in animal genomes. In plants, there has been little large-scale exploration of NAGNAG splicing, even in model plants such as Arabidopsis thaliana. To address this, here we describe the first tissue-specific database of NAGNAG motifs and their expression ratios between two splicing sites in A. thaliana. Using 435 RNA-seq datasets, we systematically identified and quantified 1,890 unique NAGNAG motifs as well as the variation of expression ratio between two splicing sites from six tissues, including 200 motifs with two novel splicing sites and 1,208 motifs containing at least one novel splicing site. Using this database, it is now possible to browse, search, and download Arabidopsis NAGNAG motifs. The collected information can also be used to retrieve the related tissue-specific transcript and amino acid information. The database largely expands our knowledge about NAGNAG events. Placing these data in a unified browsing platform should enable high throughput functional validation of the identified NAGNAG events and understand the regulatory role of NAGNAG splicing motifs through the variation of expression ratios.
 |